|
Release Date: 17 Jul 2018
The ability to replicate is the primordial definition of life. Cells propagate by making copies of themselves through replicating their DNA genome, which are blueprints of their identities. Every full grown human came from a single fertilized egg cell after approximately 10 million-billion cell divisions. New cells continue to replace old ones at a rate of over one million per second throughout life. Each cell division requires the exact replication of the genome such that each daughter cell receives a complete complement of the identical genetic information in the form of DNA. Aberrant DNA replication is the cause of many cancers and developmental disorders. Aging is also a general phenomenon of the breakdown of the replication machinery.
Unlocking the secret of DNA replication is key to understanding the mystery of life. With Watson and Crick’s determination of the DNA double helix structure in 1953, the complementarity of the two DNA strands immediately suggested how DNA is replicated. As a postdoc at Stanford, Prof Bik Tye (currently Visiting Professor of Life Science at HKUST) dedicated herself to study this fundamentally important problem. In 1984, her group at Cornell identified the proteins key to DNA replication in fungi, plants and humans; she named them mini-chromosome maintenance or MCM for short (Maine et al., 1984).
The DNA replication machine turns out to be much more complicated than imagined because of its large size (millions of atomic mass units), its multi-partite nature (consisting of three engines) and its flexibility (many movable parts). Although laboratories around the world put in Herculean efforts to tease out bits and pieces of information over the past decades, the overall picture remained murky. Up until now, the DNA replication machine has been depicted in textbooks only as cartoons with scant details (Figure 1). To put this dire situation in perspective, the Central Dogma in molecular biology is that DNA is the genetic blueprint that instructs the synthesis of tens of thousands of RNAs and proteins, the major workhorses of a cell that carry out most cellular functions (Figure 2). Deciphering the structures and therefore the functions of the machines that synthesize these macromolecules is central to molecular biology. In contrast to the RNA synthesizing machine (RNA polymerase, Nobel prize 2006) and protein synthesizing machine (ribosome, Nobel prize 2009), the DNA synthesizing machine (replisome) is still very much a black box (Figure 2).
When Tye came to UST in 2013, on the heels of the “resolution revolution” of cryogenic electron microscope (cryo-EM) technology (Nobel prize in Chemistry 2017), she decided to take advantage of this new technology to determine the atomic structure of the DNA replication machinery. Collaborating with Prof Ning Gao’s cryo-EM team in Beijing, her UST team, including two IAS Junior Fellows, Dr Yuanliang Zhai (currently Assistant Professor at the University of Hong Kong) and Dr Yongqian Zhao, published a series of articles, opening the door for deciphering the function of the DNA replication machine at unprecedented resolutions.
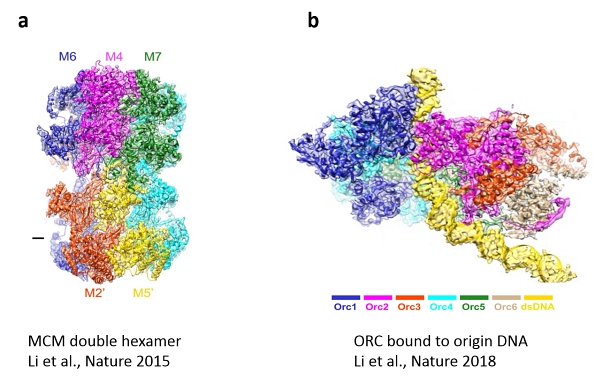
The first, published in Nature 2015, determined the structure of the MCM complex as the core engine of the DNA replication machine (Figure 3a). This breakthrough was highlighted in News and Views (accompanying commentary in same issue) marveling at the emergence of a new era for the comprehensive understanding of DNA replication. The second reported an open-ringed structure of the Cdt1-Mcm2-7 complex as a precursor of the MCM double hexamer, providing insight into the mechanics of the engine for DNA unwinding. The article in Nature this month details the atomic structure of the Origin Recognition Complex (ORC) that selects start sites genome-wide to initiate DNA replication (Figure 3b).
|
| Fig 3. CryoEM images of the MCM double hexamer (a) and ORC bound to origin DNA (b)
|
|
Structure informs function. Together, these DNA replication-related atomic images and those from other laboratories can be connected to produce a movie of the DNA replication machine in action. The door is now wide open for many more details of DNA replication to be worked out. A fundamental understanding of DNA replication at the atomic level will have an immense impact on the study of cancer, aging and other developmental disorders. The Hong Kong - Beijing team is well-positioned to continue their lead in foraging the unknown territory of DNA replication.
| 1. | Maine G, Sinha P, Tye BK. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics (1984) 106: 365–385. |
| 2. | Li N, *Zhai YL, Zhang Y, Li W, Yang M, Lei J, Tye BK, Gao N. Structure of the eukaryotic MCM complex at 3.8 Å, Nature (2015). DOI: 10.1038/nature14685 |
| 3. | Bochman M, Schwacha. DNA replication: Strand separation unraveled, Nature (2015). DOI: 10.1038/nature14643
|
| 4. | *Zhai YL, Chen E, Wu H, Li N, Yung P, Gao N, Tye BK. Open-ringed structure of the Cdt1–Mcm2–7 complex as a precursor of the MCM double hexamer, Nature Structural & Molecular Biology (2017). DOI: 10.1038/nsmb.3374
|
| 5. | *Zhai YL, Li N, Jiang H, Huang X, Gao N, Tye BK. Unique roles played by each of the non-identical MCM subunits in DNA Replication Licensing. Molecular Cell (2017) DOI: 10.1016/j.molcel.2017.06.016. Review.
|
| 6. |
Li N, Lam WH, *Zhai YL, Cheng J, Cheng E, *Zhao Y, Gao N, Tye BK. Structure of the origin recognition complex bound to DNA replication origin, Nature (2018). DOI: 10.1038/s41586-018-0293-x
|
* IAS Junior Fellow |
|


